Samuel Sternberg ’07’s latest discovery could be a giant leap in gene editing.
Columbia College | Columbia University in the City of New York
Samuel Sternberg ’07’s latest discovery could be a giant leap in gene editing.
Photographs by Jörg Meyer
What’s more, his Ph.D. advisor and co-author, and a co-founder of the biotech startup, was Jennifer Doudna, the biochemist known for her revelatory work showing how a molecule called CRISPR-Cas9 could be used to edit an organism’s genetic material; the discovery earned her a share of the Nobel Prize in chemistry last year. “I wanted to make sure I put my head down and worked on our research program, and didn’t try to rest on the laurels of Jennifer’s accolades,” Sternberg says.
Insecurities aside, Sternberg had in fact established himself as one of the country’s leading experts in CRISPR-Cas9 — with patents and prominent papers to his name — and he had ideas for expanding on the promise of that technology. His focus paid off almost immediately. Within months, Sternberg’s Columbia lab had a major paper in Nature, about so-called “jumping genes,” which could prove even better than CRISPR for gene editing. “Now we’re doing all these things that three years ago would have seemed well beyond reach,” he says. The work could someday help researchers treat cancer, produce biofuels, or make crops hardier and tastier. Indeed, last year Sternberg won a $2.4 million grant from the NIH, awarded to “especially creative” scientists working on innovative, high-impact projects.
I mentioned Sternberg’s initial self-doubts to Sanne Klompe GSAS’22, a graduate student in his lab and the lead author of the Nature paper. “That’s funny you say that,” she says. “I don’t know if he realizes how good he is.” Reflecting on her decision to join his lab, she says, “I knew he was going to be great. And I wanted to be part of that.”
Sternberg grew up in Lancaster, Pa., playing piano, saxophone and baseball. (He almost went to Oberlin for musical performance.) His dad was a geology professor, and Sternberg did science fair projects, but he was never passionate about the subject until he took chemistry and biology courses at the College. In high school bio, he’d learned about leaves and frogs, but not molecules. “It was taking organic chemistry sophomore year that I remember as being pivotal in awakening my true scientific curiosity,” he says. “Learning how chemicals can transform in very predictable ways. And that there’s a kind of logic.”

Sternberg with his Ph.D. advisor, biochemist Jennifer Doudna, and Columbia lab colleague Sanne Klompe GSAS’22.
COURTESY SAMUEL STERNBERG ’07
The other pivotal point came junior year, when he joined the lab of Ruben Gonzalez, an enthusiastic researcher “at the top of his game” who served as a great mentor. Sternberg was inspired enough to take on his own project in the lab — studying how cells complete the production of proteins — an unusual opportunity for an undergrad. “He was fantastic at the lab bench, and quickly became a voice in research meetings,” Gonzalez says. “I always told him he operated like a graduate student.” Sometimes he operated like a sailor. “His notebooks were very colorful,” Gonzalez says. “Let’s just say that when experiments weren’t working, you could tell.”
Sternberg stayed a year and a half past graduation to finish the project, becoming the lead author of a paper in Nature Structural & Molecular Biology. “Having gone through my first experience of not just how you do research at the bench,” Sternberg says, “but also how you plan experiments, how you put together a series of data into one story. at was an invaluable experience.”
Sternberg joined Doudna’s lab at UC Berkeley as a grad student in 2009, shortly before she made the discovery that would earn her a Nobel Prize. The lab had just begun working on CRISPR, which was a bit risky professionally, because at the time it was just “this very niche, esoteric thing in bacteria,” Sternberg says, and not the tool for studying cells or the therapy for treating genetic diseases that it is today.
In five years at Berkeley, including a six-month stint back at Columbia working with biochemist Eric Greene, Sternberg co-authored several important papers focusing on the Cas9 enzyme — a feature of many CRISPR systems — and how it works. One showed how to engineer versions of Cas9 that snip DNA more accurately, work that several companies have since licensed.
“When he was in the lab, I thought of him as our Renaissance man,” Doudna says. “Wonderful scientist, incredible musician, loved to have a good time and share ideas, a deep intellect, loved to debate ideas in science.” When asked for possibly embarrassing material, she mentioned his Michael Jackson tribute band. “The lab finally talked me into going to one of his performances in San Francisco, and it was a ton of fun,” she says. “I just loved it. It was a side of Sam that I hadn’t had a chance to experience.”

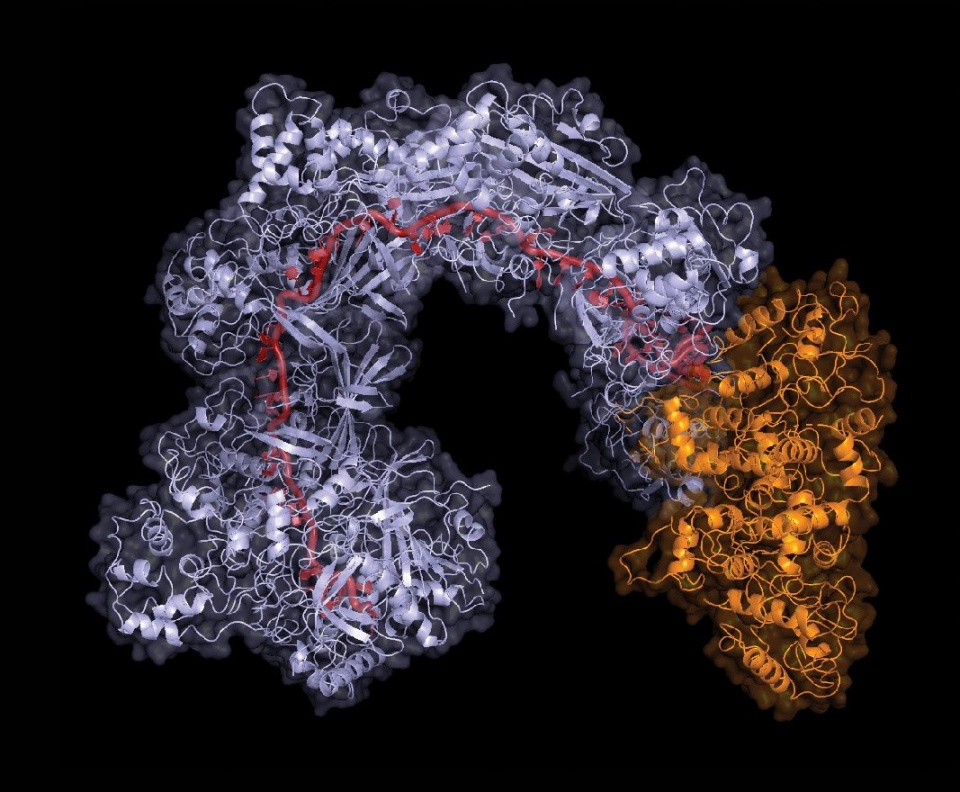
Sternberg and Klompe analyze the 3D structure of the CRISPR molecular system called Cascade (also seen below).
SAMUEL STERNBERG ’07
Sternberg worked with Doudna again at Caribou, the company she co-founded to further develop gene-editing technology; he was an early employee, acting as both a scientist and a leader of the technology development group. He learned a couple of lessons there, he says. First, he picked up the managerial professionalism he would bring to his lab at Columbia a year later. Second, he learned that he preferred academia — having the opportunity to experiment and explore ideas without concern for a bottom line.
Do jumping genes hold the key to a safer, more reliable gene-editing tool?
“The first discovery we made in my new lab is the perfect example of a project that would have been too speculative for a private company to pursue,” he says, “but has been absolutely transformative in creating new versions of CRISPR.”
One downside of CRISPR-Cas9 as a gene-editing tool is that it slices through DNA, which a cell must then repair. Many things can go awry in this process. While working at Caribou, Sternberg came across a paper on transposons, or jumping genes, which could offer an alternative. A jumping gene is actually a cluster of genes that reproduces by taking over bacterial machinery. It inserts itself into a bacterial genome and essentially forces the cell to reproduce it. Its clones then hop over to new bacterial cells and repeat the process.
Critically, a jumping gene inserts itself into a genome without creating the kind of DNA damage that CRISPR-Cas9 does. What’s more, the paper’s authors noted that some jumping genes contain the genetic code for a CRISPR system called Cascade, raising the possibility that jumping genes use it to target specific sites in bacterial genomes. Sternberg hoped to test this hypothesis in his Columbia lab (he had applied for faculty positions and been welcomed back in 2018). If jumping genes insert DNA into genomes without harming them, and do so in precise ways, they could perhaps be harnessed as a new gene-editing tool — one that would be safer and more reliable than any other at scientists’ disposal.

Sternberg and Dennis Zhang ’24 discuss data while plotting future experiments.
On our tour, Sternberg pointed out various machines that duplicate DNA or purify proteins. I watched as Klompe used a device to “heat shock” cells so that the DNA designed by the lab would enter them. “To be quite truthful, we don’t have much distinctive equipment,” Sternberg told me. “Our distinction is discovering new CRISPR systems.”
Much of that discovery happens on computers. “In the first few months in the lab, Sanne was spending most of her time at the computer sifting through genomes, deciding which candidate genes to prioritize to go after this hypothesis,” Sternberg says. On a computer in his office, he showed me the sequence of DNA letters for Cascade, alongside its 3D rendering. The cluster of proteins vaguely resembled a seahorse, with a spine made of guide RNA. Guide RNA is the genetic sequence a CRISPR system uses to target specific locations in a host genome. Wherever this bit of guide RNA matches a stretch of DNA, CRISPR does its gene-editing work.
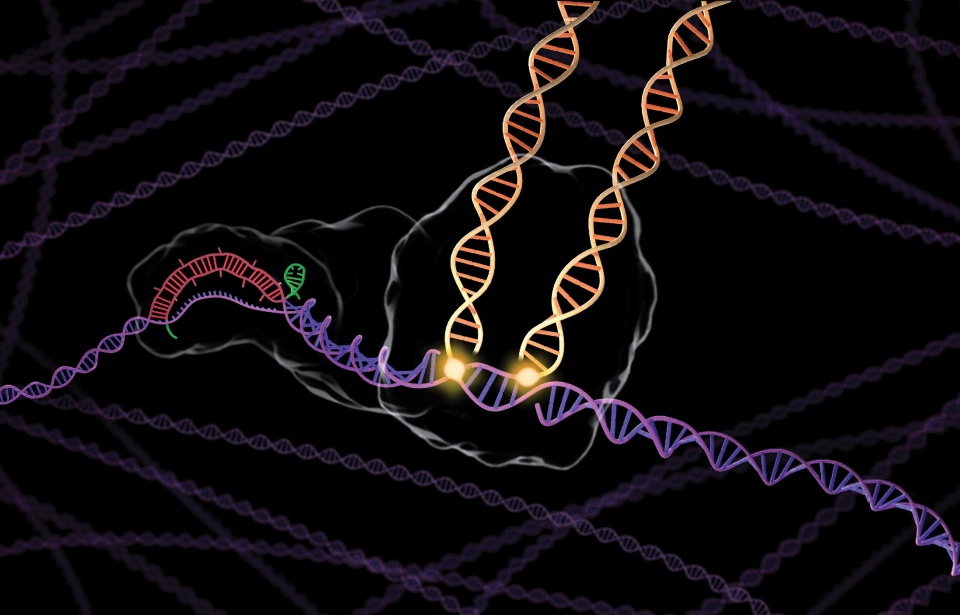
The rendering at right shows a new CRISPR technology, developed in the Sternberg Lab, that enables RNA-guided DNA integration. Unlike conventional CRISPR-Cas9 tools for gene editing, this technology inserts DNA without cutting.
MYLES MARSHALL
They ran more experiments and published the Nature paper, which, Sternberg says, “landed the lab on the map, landed Sanne’s career on the map and in a ridiculous way played out the exact way that I had hoped and dreamed when I first proposed: Hey, what about we go after this project in my new lab at Columbia?”
Sternberg’s lab — and others, too — are now expanding on that research. For example, his lab is applying the tool to human cells and is working with the Cystic Fibrosis Foundation to replace the defective gene that causes the respiratory disease. They’re working with Harris Wang, a synthetic biologist at Columbia, to engineer jumping genes that can operate in the complex environment of the gut microbiome. And they’re looking for even better versions of Cascade in the diversity of living organisms. “We can barely compete with what nature has already invented,” Klompe says.
Sternberg’s lab now stands at about 15 people. “What I really appreciate in him as a mentor is that everything is a conversation,” Klompe says. “Instead of telling me what to do, it’s almost like, OK, what shall we do next?” When the lab was small and people stayed late to work on that first paper, Sternberg would order dinner. “It was like a little scientific family having dinner in the break room,” she says. The lab’s walls feature fliers celebrating members’ achievements; one advertises a lab outing to the sci-fi movie Rampage, in which a CRISPR-modified pathogen leads to the creation of a giant gorilla, wolf and crocodile. According to Sternberg, “We built an environment that’s not just about the science, but also a place where people will grow as individuals and develop their career aspirations.”
Colleagues have not been surprised to see Sternberg’s success. “His scientific instinct is amazing,” Ruben Gonzalez says. “It’s been a lot of fun to watch.” (The two still get beers and are collaborating on a project.) According to Doudna, “He’s doing some of the most creative work in the discovery of CRISPR systems right now and figuring out how they work.” Walter Isaacson, who wrote the best-selling biography The Code Breaker: Jennifer Doudna, Gene Editing, and the Future of the Human Race, met with Sternberg regularly to pick his brain. “There’s a boyish enthusiasm,” Isaacson says. “He’s excited about the smallest details of how nature works.”
Sternberg still sometimes questions why he’s the one with an office. He notes that some members of his lab have expertise surpassing his own. “My biggest learning experience here has been realizing that that’s actually OK,” he says. “Being a good scientist doesn’t mean you know how to do everything. One of the best skills I learned from Ruben and from Jennifer is to be resourceful, and to engage with your colleagues. It’s all about putting all the different skills in the pot.”
Matthew Hutson is a freelance science reporter in New York City and a contributing writer at The New Yorker.

Published three times a year by Columbia College for alumni, students, faculty, parents and friends.
Columbia Alumni Center
622 W. 113th St., MC 4530, 6th Fl.
New York, NY 10025
212-851-7852
cct@columbia.edu

Columbia Alumni Center
622 W. 113th St., MC 4530, 4th Fl.
New York, NY 10025
212-851-7488
ccalumni@columbia.edu